Schwaggy P's Random Stuff
- Thread starter Schwaggy P
- Start date
H.A.F.
a.k.a. Rusty Nails
I had to take chemistry and biology in college, and have been looking at some of the old textbooks. I think the simplest breakdown of this is that the trait is there, even if you don't see it in that particular plant.We have an OG Kush female we kept from a pack of fems. This OG did not express any intersex traits while growing, but some of her sisters did. We feel good about this plant because the phenotype did not show any hermie trait, but let’s remember we cannot see the genotype.
In the example above if there were 1 of 4 plants that did not show intersex traits, that doesn't tell me to use that plant as a mother. It tells me that the strain has a 75% chance of herming. Shitcan it. If there's a trait you really want there, go back a step in the lineage breed a non-herming version for that part of your intended pairing.
I have never grown a male plant out, but I can understand basic concepts. I would correlate this to the issues I have heard about breeders selling seeds, and knowing that they might herm. They skipped a step IMHO.
yougrowyourway
illgrowmyway
Not an opinion, that's a fact. They rather have the quick buck. IMO people today rather have a variety with their popping. Imagine working a line for even a year or two then go to move it and noone even bats an eye at it. Ofcourse if it's true fire it will eventually catch on but at what cost. How long? If this isn't a side hobby for you that's a loss and a half. They skipped a step IMHO.
gwheels
Hobby Farmer
First of all....thanks again man. Skunky VA is Stupendous.... Monster fat dank fruity skunky hashy wonderful buds. One pair left to chop. Day 65...

Secondly thanks for taking the time to make these articles. I find it really interesting. After really thinking about it after reading the latest articles about female male traits, I culled my 2 males today.
Thats enough of that man. I have fire seeds.
And thats a lot of thinking...that goes into good genetic seeds. I have enough other stuff to get right without dipping my toe where it doesnt belong. And i just know what is going to happen.
Bhangi Haze X.....everything in the bloom room seeds because it stuck to my sock or something.
Or even worse i get seeds....plant them and end up with a 10 foot tall sativa that produces 2 oz of the most average plain buds you ever grew ! so i will take a hard pass !
A mans got to know his limitations (The Outlaw Josey Wales!)
I have 3 more skunky VA in veg. I will practice growing

Secondly thanks for taking the time to make these articles. I find it really interesting. After really thinking about it after reading the latest articles about female male traits, I culled my 2 males today.
Thats enough of that man. I have fire seeds.
And thats a lot of thinking...that goes into good genetic seeds. I have enough other stuff to get right without dipping my toe where it doesnt belong. And i just know what is going to happen.
Bhangi Haze X.....everything in the bloom room seeds because it stuck to my sock or something.
Or even worse i get seeds....plant them and end up with a 10 foot tall sativa that produces 2 oz of the most average plain buds you ever grew ! so i will take a hard pass !
A mans got to know his limitations (The Outlaw Josey Wales!)
I have 3 more skunky VA in veg. I will practice growing
Last edited:
yougrowyourway
illgrowmyway
I'm right there with you. However with much thought. I've got my own lil creation I'm looking to create that I'm willing to dedicate years to. All on the side of my regular garden. But that's prolly a lil down the road from now anyway.I have enough other stuff to get right without dipping my toe where it doesnt belong. And i just know what is going to happen.
A mans got to know his limitations (The Outlaw Josey Wales!)
I have 3 more skunky VA in veg. I will practice growing
Schwaggy P thx for the biology lesson! I have a few questions. How do we know which cannabis traits are polygenic and/or multi-allele? How would find out during breeding?
What happens to the Punnett Squares when you have polygenic traits in cannabis. How about multi-allele traits in cannabis? What happens to the Punnett Squares when you have both polygenic and multi-allele traits?
Of these cannabis polygenic and multi-allele traits, how do we know which are dominant and which are not?
How do we know which cannabis strains carry polygenic or multi-allele dominance and in which phenotypes?
How do polygenic and multi-allele trats, as well as the lack of genomic data, specifically limit targeted cannabis breeding projects?
Is the cannabis hermaphroditism trait polygenic, multi-allele, a combination, or caused by some other mechanism or combination of mechanisms? If something else, what?
Without having the cannabis genomic data or knowledge of its polygenic and multi-allele traits, how do you true breed?
How does the science of epigenetics relate to breeding cannabis?
What happens to the Punnett Squares when you have polygenic traits in cannabis. How about multi-allele traits in cannabis? What happens to the Punnett Squares when you have both polygenic and multi-allele traits?
Of these cannabis polygenic and multi-allele traits, how do we know which are dominant and which are not?
How do we know which cannabis strains carry polygenic or multi-allele dominance and in which phenotypes?
How do polygenic and multi-allele trats, as well as the lack of genomic data, specifically limit targeted cannabis breeding projects?
Is the cannabis hermaphroditism trait polygenic, multi-allele, a combination, or caused by some other mechanism or combination of mechanisms? If something else, what?
Without having the cannabis genomic data or knowledge of its polygenic and multi-allele traits, how do you true breed?
How does the science of epigenetics relate to breeding cannabis?
Thank you for the updates and I'm very glad you're pleased with the Skunky VA!First of all....thanks again man. Skunky VA is Stupendous.... Monster fat dank fruity skunky hashy wonderful buds.
My pleasure!Schwaggy P thx for the biology lesson!
So the reason I’d keep things simplified to the Mendelian genetics is the practical limitations on home breeding. Without gene markers and a genomics lab, there’s no real way for the home breeder to be sure. You can design small scale breeding experiments to benchmark a specific plant for one-two traits and observe the trait through generations. By looking at the frequencies of the trait’s expression and comparing them to the frequencies expected for different genotype arrangements, you could make decent assumptions about your target plant’s genetic makeup for the trait.How do we know which cannabis traits are polygenic and/or multi-allele? How would find out during breeding?
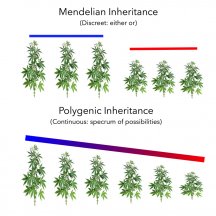
The practical difference between the discreet inheritance vs. polygenic can be expressed in an example. Let’s assume our trait of interest is height. In our simplified Punnett Square, the plants will either be “tall” (Hh), (HH), or “short” (hh). This would imply that there are only 2 possible final heights our plants could express.
If height is dictated by the dance of multiple genetic contributors (polygenic factors), then there is a range of possibilities in the expression of height. This manifests as a population with varied height that makes delineation between “tall” and “short” to be a threshold decision. I made this illustration to show this difference:
Something to keep in mind: while these concepts are presented in their purest theoretical form, the practical manifestation is rarely one or the other extreme model. When popping a pack of seeds, it’s not shocking to observe the plants’ height to take on a “gray” area between the two models presented in the illustration. You’ll find that when focused on height, you see not all the same tall and/or short, nor do you see a perfect crescendo of increasing/decreasing height. You may observe a tall/medium/short arrangement of heights such that they are discreet heights of a spectral nature.
The presence of polygenic traits does not mean that the final expression of said trait is dictated by the contributing factors evenly. For example, if 12 different genetic contributors code height, there’s no reason to assume all 12 contribute equal influence, there could still be one majority-contributing factor with 11 slightly nuanced contributors. This uneven distribution of influence would allow one, in effect, to treat the trait as a discreet inheritance situation.
Understanding these pure theoretical models of inheritance for practical breeding programs allows one to color their expectation and hone their ability to make the selections, which will best suite their goals. When you set out to breed a short plant, it’s doubtful you have an exact height in mind, so for purposes of Punnett Square use, you could evaluate the shorter plants as “short” and continue with a discreet inheritance model until your results fall outside of the acceptable range of tolerance for the trait you’re after. At which point you can assume the trait is of a polygenic nature expressing outside your ability to predict.
The Punnett Square ceases to be useful with polygenic traits (assuming you'd like to illustrate them). The nature of polygenic traits means that the assumption of discreet inheritance (Mendel’s model) fails, as you are now dealing with a spectrum of variation. This variation as a function of the number and degree of contribution to the trait of interest leaves the assumptions of the Punnett Square incapable of giving a practical outlook on expectations.What happens to the Punnett Squares when you have polygenic traits in cannabis. How about multi-allele traits in cannabis?
Think about the illustration above and how one would prepare a Punnett square that includes every possible combination of every height possible. Now try to expand the Punnett Square to include every possible combination of multiple traits and the square stops being a practical tool for at-home breeding.
The Punnett Square would have to be severely amended to include distribution curves relating to the spectrum of variation. At this point it stops being a Punnett Square and looks more like a collection of distribution graphs.What happens to the Punnett Squares when you have both polygenic and multi-allele traits?
You couldn’t know for sure. At best you can conclude the contributing genetic factors are such that no pattern can be discerned, or that the contributions can be assessed as inconsequential and adopt a discreet model of inheritance (example provided later).Of these cannabis polygenic and multi-allele traits, how do we know which are dominant and which are not?
How do we know which cannabis strains carry polygenic or multi-allele dominance and in which phenotypes?
Without a genomics lab, observation and careful note taking. If you observe that a particular trait of interest is not following a predictable “discreet” frequency of expression in subsequent generations, then you may well assume the trait is polygenic. How you would then treat this trait depends on whether your plant numbers are sufficient enough to discern the potential expressions of this trait.How do polygenic and multi-allele trats, as well as the lack of genomic data, specifically limit targeted cannabis breeding projects?
For practical purposes, I have never come across a breeder that is either expecting to be able to, much less putting to work, the ability to guarantee specific genotype stability in each seed. Meaning, it’s not realistic to expect to be able to breed a line so true that you could guarantee the exact height/potency/flowering time, etc. to the inch/%/day etc. You are dealing with a range of variation. It would be realistic to expect to produce a line of specific traits that express within a range of “acceptable” manifestation. So while I can’t say that every seed will result in a female finished at 63.4days at 2.36’ with 18.23% THC; I could achieve my goals to be able to say that this seed will finish around 60-67days, 2-2.5’, with 16-19%.
I can say I have this Afghan Skunk line that is short. I can’t tell you the exact height, but through selection and testing, when flipped at 1.5’ the progeny will finish between 2-2.5’. The deciding factor between one plant finishing at 2’ and another at 2.5’ may very well be polygenic, but the genotypes have been trimmed to be within this range of possibility, so I can treat the trait as discreet. My treating the trait as discreet isn’t ignoring the fact that it’s polygenic; it’s simply an adoption of a practically useful tool with the understanding that, as with most scientific models, this is an approximation.
At best, I could say that in a pack of seeds, you’ll find a short plant of high potency and quick maturation, but could not guarantee that every seed will express an exact phenotype.
I would speculate that all traits are polygenic. The question is to what extent is the trait dominated by variable contributing factors. Is the resulting spectrum of possibility such that it can be treated as discreet within a set range of acceptance? What of co-dominant contributions resulting in a “new” trait (think red flower and white flower creating pink flowers)? I’d familiarize myself with the possibilities and allow the manifestation of these traits through observation guide which assumptions to accept for further work.Is the cannabis hermaphroditism trait polygenic, multi-allele, a combination, or caused by some other mechanism or combination of mechanisms? If something else, what?
No two genotypes are the exact same, and I would have to assess each plant independent when it comes to questions of specific traits and how they express/breed.
Here is an example of a far better equipped group exploring this concept:
Even in a high-tech environment, the assumption of simple Mendelian inheritance is a useful model (red). While the hermie trait is in fact a polygenic trait gaining it’s form through the contribution of many genetic affecters, the expression is discreet (blue). Were I breeding plants that behaved exactly like this, my limited inheritance model (discreet) would still work, regardless of my ignorance of the far more intricate underlying polygenic nature. In essence, the polygenetic traits are irrelevant if the final expression takes on a discreet form.
This could be a long response, so I’ll limit it to the overall approach:Without having the cannabis genomic data or knowledge of its polygenic and multi-allele traits, how do you true breed?
Assume a discreet model (as long as the range of trait expression is within your acceptance) and take notes about your frequencies. Keep your traits limited, observe their frequency, compare to the Punnett predictions to “reverse engineer” the genotype. Then move forward with another trait. In short, you’d have to benchmark your plants at each step.
Epigenetics relates to the phenomena of transcendent genetic affects relating to functional groups (methylation, phosphorylation, etc.) attaching to DNA affecting transcription and gene expression.How does the science of epigenetics relate to breeding cannabis?
Certain chemical functional group molecules can attach themselves to the DNA and histones (proteins DNA wrap around). These attached groups can affect how the DNA is wrapped affectively “shutting off” certain gene expressions. These can come from environmental factors, stress by-products, chemical additives, etc. and affect the genetics in such a way as to alter their original potential expression.
I’ll use an analogy to simplify this. Let’s assume I have my favorite album on CD and I listen to it all the time. One day, I accidentally drop the CD on the floor and lightly scratch on part of the CD. Every time I play the CD, when the laser gets to this small scratch, the music skips. If I try to copy this CD or play it, I’ll forever have this skipped part of the music. Does this mean the album has been so destroyed that I couldn’t tell it’s still my favorite band? Nope, it just skips that little bit of the guitar solo on track #3.
The DNA is like the CD in that it contains all of the information I need to “replay” this set of info. If epigenetic factors “scratch” one segment of this info, I will now experience a small imperfection moving forward with genes being “lost” and will now affect any copies of the information being made in the future.
I would speculate that while the epigenetic alterations to genetic material could result in large observable changes in a population, they’d mostly take the form of very low-level disturbances. So I would be more apt to expect one vein pattern of one leaf blade to be slightly more fractal than otherwise expressed, more so than the plant now producing 27 blade leaves.
To the extent some claim cuts are experiencing “genetic drift” as they are kept in different environments for periods of time, I’d credit epigenetic factors. I don’t really buy the claim that a cut has taken on the exaggerated differences some claim as genetics are more resilient than a few years in different mother rooms could impress.
I am not aware of any specific procedural points to be aware of during cannabis breeding in order to avoid unwanted epigenetic factors playing a role. There are some genetic modifications you can introduce in order to induce minor/severe genetic manipulation. These techniques are widely used in agricultural breeding and involve purposeful mutations through chemical means in order to alter a plant’s genetics. I’ve played around with this idea but don’t post much about it as many have differing opinions and presuppositions about GMO breeding and don’t feel like having to get mired in thinly veiled political debates about stuff no one else is even doing. I don’t use it on any released work (so don’t worry if you’re growing Schwaggy Seeds).
An example of this that many may be familiar with is GA3 used early to induce exaggerated growth rates. This technique is temporary and is regulated by the presence of the chemical stimulant. There are other methods of permanently altering the DNA of plants in order for the mutation to be heritable (I’ve messed with radioactive smoke detector material (Americium-241) to introduce limited ionizing radiation to plants), but is very hit/miss as far as at-home practical use. It’s something to be aware of, but I wouldn’t worry too much as I’m of the opinion that these issues at best dance on the periphery of genetic alterations.
gwheels
Hobby Farmer
Like a cartoon !Thank you for the updates and I'm very glad you're pleased with the Skunky VA!
Attachments
-
88.8 KB Views: 36
GCG
Dinosaur
Holy hell, Im with @gwheels. May attempt some personal chucking but will never call it breeding and will be strictly for play. Realize all my breeding dreams were more, I hope I get lucky dreams. Way more goes into creating stable genetics than I have the intellect, time or space to provide. Not sure if that's a good or bad realization but it is reality.
@Schwaggy P is it possible for big AG and their unlimited resources to create and market seeds that will hit specific markers? Color, height, potency or will it always be a range. Have to assume it's in their plans if its theoretically possible.
@Schwaggy P is it possible for big AG and their unlimited resources to create and market seeds that will hit specific markers? Color, height, potency or will it always be a range. Have to assume it's in their plans if its theoretically possible.
Well if it helps, think of some of the most popular strains (Chemdog, GSC, Sour D, OG Kush) and realize these were hermie accidents and not the result of dedicated selective breeding over many years. Pretty funny when you think about it.Holy hell, Im with @gwheels. May attempt some personal chucking but will never call it breeding and will be strictly for play. Realize all my breeding dreams were more, I hope I get lucky dreams. Way more goes into creating stable genetics than I have the intellect, time or space to provide. Not sure if that's a good or bad realization but it is reality.
I’d say they have the ability to make seeds that are “practically” hitting the mark. But I wouldn’t think they’d be after breeding for seed making. Think about tobacco, there are no large scale tobacco breeding for seed markets where people are home-growing tobacco for cigarettes. The industry would be more geared toward large scale flower production bringing the per unit price so low as to make home growing for the average smoker more expensive and time consuming to bother with.@Schwaggy P is it possible for big AG and their unlimited resources to create and market seeds that will hit specific markers? Color, height, potency or will it always be a range. Have to assume it's in their plans if its theoretically possible.
H.A.F.
a.k.a. Rusty Nails
Bingo. But people still brew beer at home, or pay more for micro-brews.The industry would be more geared toward large scale flower production bringing the per unit price so low as to make home growing for the average smoker more expensive and time consuming to bother with.
Mine has bulked up immensely, but mine is granny VA, lol. Craziest plant I seen in a while. Where's the smell? Lol. I did see skunkva or pbud or someone on IG post a pic of skunk and he was saying white pistils matured, bulky, which is like mine. I'm chopping in a day or two, I'll throw a pic upLike a cartoon !Skunky VA can pack on some mass.
Baja.Beaches
Super Active Member
I am back from my extended travels, I brought plenty of Skunky VA & smoked it everyday with friends. That strain was loved by all, the taste, loud smell, & bag appeal all stellar. Outstanding in every way. 
I would have loved to have brought the one pheno of the Granny Skunk cross too for a side by side but it was not dry enough at the time. It has cured wonderfully while I was gone, still extremely loud old school skunk style, Outstanding as well, I am loving it. Sounds like @Bodyne had very different results indoors. Interesting.
Smoke comparison week for me finally. Fun stuff. Do you have a smoke report form that you like?
I would have loved to have brought the one pheno of the Granny Skunk cross too for a side by side but it was not dry enough at the time. It has cured wonderfully while I was gone, still extremely loud old school skunk style, Outstanding as well, I am loving it. Sounds like @Bodyne had very different results indoors. Interesting.
Smoke comparison week for me finally. Fun stuff. Do you have a smoke report form that you like?
here's mine, as soon as it dries out a bit, im choppin it. Bulked up bigtime last week or so. Still not much smell, lol.
Attachments
-
186.4 KB Views: 50
Welcome back! Feel free to describe them however you'd like.I am back from my extended travels, I brought plenty of Skunky VA & smoked it everyday with friends. That strain was loved by all, the taste, loud smell, & bag appeal all stellar. Outstanding in every way.
I would have loved to have brought the one pheno of the Granny Skunk cross too for a side by side but it was not dry enough at the time. It has cured wonderfully while I was gone, still extremely loud old school skunk style, Outstanding as well, I am loving it. Sounds like @Bodyne had very different results indoors. Interesting.
Smoke comparison week for me finally. Fun stuff. Do you have a smoke report form that you like?
She has really blown up.here's mine, as soon as it dries out a bit, im choppin it. Bulked up bigtime last week or so. Still not much smell, lol.